1 - Introduction to the SampleData data platform¶
This first User Guide page/notebook will:
present the main features of the data format associated with the platform
list the prerequisite knowledge that we recommend to master before starting to use SampleData
Note: This page/notebook does not include Python code to learn how to manipulate the SampleData class. However, it provides a complete overview of the scientific challenges that motivated the implementation of the class, which surely helps gaining insight in using the code and understand relevant applications.
I - The SampleData platform¶
The pymicro package is used on a daily basis by material scientists teams involved in the development of the data paradigm presented in section I. Its original design, that relied on classical Python and Numpy data objects to handle material data, dit not allow to efficiently handle the multi-modal 4D material science data challenges. Therefore, an new subpackage has been introduced within pymicro, the pymicro.core subpackage, to serve as pymicro’s data platform.
This pymicro.core package has been designed to be independent of the rest of the pymicro code. Its aim is to serve as a prototype implementation of what could constitute a large-scale data platform the material microstructure science community. In the future, it is likely that it will become a fully independent package, and hence an external pymicro dependency. In that case, it will most likely be name SampleData, as its main class object.
SampleData is a Python class designed to implement two essential functions for the management of multimodal material data sets within the Pymicro code: 1. A unified file format and data model able to handle all data modalities presented below, within the same dataset. 2. A high-level interface allowing interactions with datasets for user and external numerical tools while hiding all the data management technical complexity
As its name suggests, it is designed to associate a dataset file for each material sample, whose data must be collected, stored and/or processed. The class then behave as a high-level interface allowing to interact with the data. Its main functionalities are: * Add, remove, and organize easily data of various types, formats, shapes into the same dataset * Add, remove and organize easily light metadata for all elements of a dataset * get simple or detailed information on the dataset content *
allow easy visualization of spatially organized data (images, maps, measured or simulated fields) * allow a flexible and efficient compression of each element of a dataset * a framework to automate interface between the datasets and data processing or numerical simulation tools * a framework to derive classes from SampleData that are specialized to a specific type of material samples, such as the Microstructure class of the pymicro.crystal.microstructure module, dedicated to
polycrystalline samples * ensure convergence of spatially organized data
Regarding this last point, the “convergence” of data refers to the conversion into a unified data model of all data representing microstructure geometries, of mechanical fields (measured or simulated), of different origins. This allows to directly visualize and process them together with the same tools.
The SampleData class is thus the interface that the data platform user has to manipulate in order to access those functionalities, and interact with the dataset. Its detailed presentation is the subject of the series of notebooks of which this document is only the first. The rest of the section is dedicated to the presentation of the file format and data model that is implement by the SampleData class.
The SampleData file format¶
Even if its implementation does not require it, the SampleData class has been designed to be used with the following convention: 1 dataset per material sample. Each dataset is associated with a pair of files:
a HDF5 file
a XDMF file
HDF5 is a library and file format that supports an unlimited variety of datatypes, and is designed for flexible and efficient I/O and for high volume and complex data. It is a hierarchical file format in the sense that it allows to gather multiple data within a file and create links to organize them in relation to each other, creating an internal file hierarchy, replicating a file system with directories and files within one file. HDF5 stores data with a binary encoding, and is compatible with many of the most powerfull compression algorithms available. It also allows to add text metadata to all data elements in the file, called HDF5 attributes. The HDF5 file, within the SampleData file pair, is the actual data set: it contains all data and metadata related to the material sample.
XMDF is an extensible Data Model to exchange scientific data between High Performance Computing codes and numerical tools. It has been practically designed for spatially organized data, i.e. fields, used as inputs and outputs by simulation tools. Practically, it is an XML file containing the description, for each grid supporting the fields, of its topology, its geometry, its dimensions, as well as the data arrays, types and dimensions of field defined on the grid. Rather than writing the heavy data describing gris and fields in to the XML file, the XDMF syntax allow to refer to arrays stored in HDF5 data files. In that case, and in the case of the ``SampleData`` file pair, the XDMF file can be seen as an external metadata file allowing to interpret raw binary data stored in a HDF5 file as spatially organized data.
One important feature of thos HDF5/XDMF file pairs, is that they are a rather standard format. In particular, a reader for this format is implemented in the powerfull and popular 3D visualization software Paraview. The ``SampleData`` class ensures that both HDF5 and XML are always synchronized together and with the content of the dataset. This means that whenever users add spatially organized data into a SampleData HDF5 dataset, they can simply visualize their dataset content by opening
with Paraview the XMDF file.
The SampleData Data Model¶
In addition to the HDF5/XDMF file pair, SampleData datasets comply with a specific data model. This section provides an quick overview of the different elements of this data model. Each of these elements will be extensively detailed in the next Notebooks of this User Guide.
This data model is built on top of the HDF5 data model, whose primary objects are:
Groups: a data structure that can be linked to other groups or datasets, and is used to organized data objects. They can been seen as the ‘directories’ of a HDF5 dataset. Every HDF5 file contains a root group that can contain other groups.
Data arrays or Nodes: arrays of data that can have different types/shapes, and are attached to a Group.
Attributes: Name/Value pairs that can take any form as long as they remain small. It is the official way to store metadata into HDF5 files. Both Groups and Datasets can hold as many Attributes as required.
The SampleData data model introduces two types of particular HDF5 Groups. They are dedicated to the representation of spatially organized data. Such data consist in geometrical grid supporting fields. These grids can be regular gris, or have more complex topologies. Those two situations correspond to the 2 Group types in the data model, that are:
Image Groups are HDF5 groups designed to store data describing 2D or 3D images, i.e. regular grids supporting scalar or tensorial fields. They are used for instance, to store data coming from SEM/EBSD or X-ray tomography imaging, or FFT simulation results. They are actually 3 types of image groups:
2DImagegroups: represent two dimensional images, i.e. grids of \((N_x,N_y)\) pixels3DImagegroups: represent three dimensional images, i.e. grids of \((N_x,N_y, N_z)\) voxelsemptyImagegroups: represent image groups that do not yet support any data and topology yet
Mesh Groups are HDF5 groups designed to store data describing 2D or 3D meshes, i.e. grids described by node coordinates and elements connectivities, supporting scalar or tensorial fields. They can be used to store data coming from finite element simulations, CAD designs etc…They are actually 3 types of image groups:
2DMeshgroups: represent two dimensional meshes, i.e. grid of nodes defined by their \((X_i,Y_i)\) coordinate pairs3DMeshgroups: represent three dimensional meshes, i.e. grid of nodes defined by their \((X_i,Y_i,Z_i)\) coordinate pairsemptyMeshgroups: represent mesh groups that do not yet support any data and topology yet
In addition, the data model introduces two additional types of HDF5 Nodes:
Structured Tables are heterogeneous and bidimensional data arrays, i.e. that may contain data of different types (integers, floats, strings, sub arrays…) within the same row, all rows having the same format. Each column of those arrays can have a specific name. Those arrays are the in-memory equivalent of Numpy structured arrays
Fields are specific data arrays that must belong to a grid group (Image or Mesh Group), and whose shape and dimensions must comply with the grid topology. They are used to store and manipulate spatially organized arrays, that represent mechanical fields (for instance, displacement or temperature fields, microstructure phase maps, EBSD orientation maps….)
Those specific data objects introduce by SampleData all have a specific data model, associated metadata, and a specific interface. They are all reviewed in details by a specific tutorial Notebook of this user guide.
Data Model example¶
To illustrate what SampleData datasets may look like, a virtual example of a polycrystalline material sample dataset is represented in the schematic diagram below:
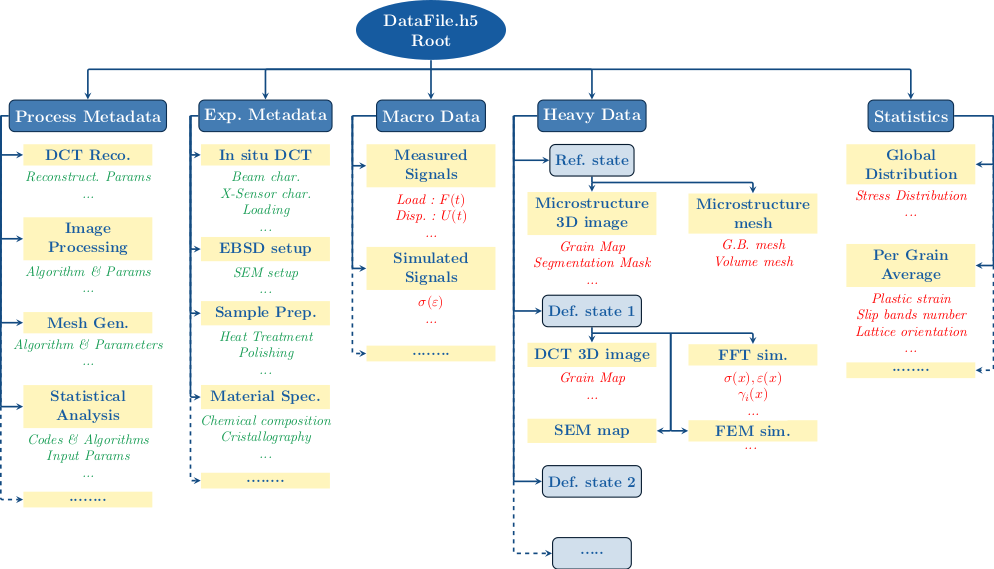
Groups are used to organize data into coherent categories. Metadata are represented in green, and datasets in red. As it appears on the diagram, Groups can be dedicated only to organize metadata, for instance to document material nature, composition, elaboration process, or the experimental set-up used for imaging and mechanical tests. The Macro Data and Statistics Groups will typically contain simple arrays datasets or structured tables, to store mechanical tests macroscopic outputs (loading curves) or some statistics on the microstructure geometry or mechanical state. The Heavy Data group will typically contain the spatially organized data, coming from 3D or 2D in-situ imaging techniques, and from numerical simulation softwares. It will most likely contain Image or Mesh Groups, filled with Fields datasets.
II - SampleData dependencies and pre-requisites¶
As SampleData is a rather complex class, relying on several libraries and packages, it is strongly adviced to take a look at and learn the basics of the following tools/packages:
Obviously, basic notions of Python (Python syntax, data types, namespaces, imports, classes, working with class methods, Python interpreters)
SampleData HDF5 interface is based on the Python package Pytables. It handles the management of the HDF5 file and the compression of datasets. Knowing its basics can be a valuable help when using SampleData.
Warning: In the series of notebooks that make up this SampleData user guide, a minimal knowledge of these elements will be assumed.
Now that the relevant context and description of the data platform has been set, you can start to learn how to use it with the second Notebook of this user guide !